

3rd Annual Multi-Omics & Microbiome
Virtual Conference
June 11-12, 2024
We are combining Multi-Omics and Microbiome R&D, Informatics, and Technology into one Virtual Conference. There will be speakers from Industry, Academia, and Government where you can see the latest and greatest accomplishments and offerings.
The last year has seen a lot of progress in integrating multi-omics. Examples include scGPT, a foundational model for single cell multi-omics, to integrating scRNAseq with human population genetics by the Teichman lab. Meanwhile new sequencing technologies and databases are helping to decipher the microbiome.
Organizations Speaking














NATIONAL INSTITUTE OF
STANDARDS & TECHNOLOGY


AGENDA

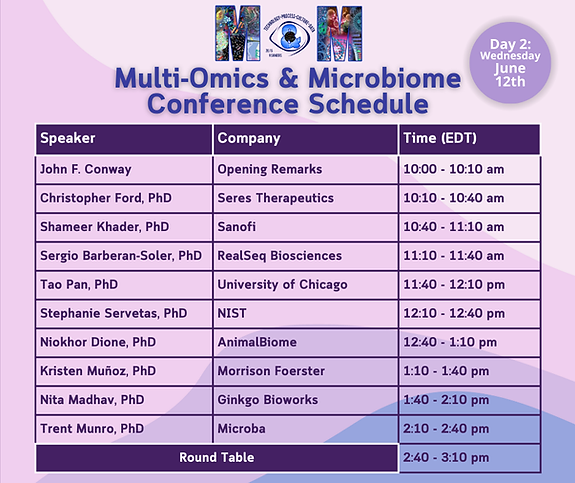.png)
SPEAKERS

Talk Title:
Scaling Drug Target Discovery using Multi-modal Evidence and Artificial Intelligence
Abstract:
Target discovery and credentialing is a critical step in the design, development and repurposing of therapeutic assets. In this talk, I will briefly discuss our efforts in combining multi-modal biological data compiled from diverse biomolecular profiling experiments and computational assessments to build Sanofi Disease Data Ecosystem. Further, I will discuss our learnings from two target discovery projects NeuroID and OcularID aim to find novel targets for neurodegenerative (Parkinson’s disease) and ocular (age-related macular degeneration) diseases. Data types including genomics, single-cell and bulk transcriptomics, proteomics, expression quantitative trait loci, epigenetic, CRISPR, pathway and clinical correlates with a suite of artificial intelligence-based scoring for the discovery and prioritization of targets.

Talk Title:
Regional Microbiota Mismatches from Fecal Microbiota Transplants Promote Persistent, Off-Target Consequences to the Host
Abstract:
Fecal microbiota transplant (FMT) is an increasingly used intervention, but its suitability to restore regional gut microbiota particularly in the small bowel (SB) must be questioned because of its predominant anaerobic composition. In human subjects receiving FMT by upper endoscopy, duodenal engraftment of anaerobes was observed after 4-weeks. We hypothesized that peroral FMT create host-microbe mismatches that impact SB homeostasis. To test this, antibiotic-treated SPF (specific pathogen-free) mice were given jejunal, cecal, or fecal-MTs (JMT, CMT, FMT) and studied 1- or 3-months later. JMT and FMT altered regional microbiota membership and function, energy balance, and intestinal and hepatic transcriptomes; JMT favored host metabolic and FMT immune pathways. MTs drove regional intestinal identity (Gata4, Gata6, Satb2) and downstream differentiation markers. RNAseq of metabolite-exposed human enteroids and duodenal biopsies post-FMT confirmed transcriptional changes in mice. Thus, regional microbial mismatches after FMTs can lead to unintended consequences and requires rethinking of microbiome-based interventions.

Talk Title:
An Advanced AI/ML Approach to Development of Precision Live Biotherapeutics
Abstract:
We have developed the worlds most advanced bioinformatic pipeline for metagenomic microbiome analysis and established a globally unique, comprehensive proprietary Microbiome databank with more than 20,000 samples matched to extensive metadata allowing us to take a human first, data driven approach to drug development. Microba deploys trained deep learning neural networks (DNN) to interrogate the host-microbial cohorts for feature importance. These DNN models identify core characteristics that discriminate between the cohorts. Our pipeline includes a clinical stage candidate for Inflammatory Bowel Disease (Ulcerative Colitis), a pre-clinical program in oncology and a discovery stage program partnered with Ginkgo Bioworks in Autoimmune Disease.

Talk Title:
AI Toolkit For Precision Oncology
Abstract:
The emergence of artificial intelligence (AI) is transforming various fields, including cancer care. This work introduces an AI toolkit designed to enhance the oncology drug development process, from target discovery to personalized treatment selection.
The toolkit supports diverse oncology processes such as target discovery, drug discovery, biomarker identification, and personalized treatment strategies. It employs machine learning models validated with CRISPR screening for target discovery, ensuring practical and actionable results. In drug discovery, state-of-the-art generative models streamline the traditionally lengthy process, accelerating the transition from concept to clinical application.
For biomarker discovery, the toolkit integrates multi-omics data with deep learning-based augmentation techniques, extracting valuable insights from small-scale clinical trials to reliably detect treatment response or resistance markers. This has been benchmarked against standard methods and deployed in actual clinical trials.
Additionally, large language models (LLMs) assist in matching patients with suitable therapies and clinical trials, addressing the need for an efficient system to connect patients with optimal treatments, thus enhancing cancer care efficacy.
In summary, this AI toolkit exemplifies the integration of AI in oncology, improving the precision and effectiveness of cancer treatment. By tackling key challenges in oncology drug development and treatment, it advances personalized medicine, leading to better outcomes and quality of life for patients. This toolkit is revolutionizing oncology drug development and reinforcing AI's role in cancer treatment advancements.

Talk Title:
Unlocking Prognostic and Therapeutic Biomarkers in Recurrent/Metastatic Head and Neck Squamous Cell Carcinoma (HNSCC)
Abstract:
HNSCC arises primarily from the mucosal epithelium of the oral cavity, pharynx, and larynx, constituting a significant portion of head and neck cancers. Ranked as the sixth most prevalent cancer globally, HNSCC is linked to risk factors such as alcohol consumption, smoking, and viral infections. Despite representing a heterogeneous spectrum of diseases, HNSCC poses substantial challenges due to its high rates of recurrence and metastasis, affecting around 20% of early-stage and 50% of locally advanced patients. While immune checkpoint blockade (ICB) therapies have demonstrated efficacy in HNSCC, a considerable portion of patients do not experience significant clinical benefits and face adverse effects. Established biomarkers like tumor mutational burden (TMB) exhibit limited predictive value in HNSCC. Tertiary lymphoid structures (TLS) have emerged as potential predictors of response to ICB therapy in various cancers, including non-small cell lung carcinoma, bladder cancer, sarcomas, and melanoma. However, the relationship between TLS presence and response to ICB treatment in HNSCC remains poorly understood.

Talk Title:
RNA Fragmentomics: Next-Generation Analytes for Accurate Microbiome Detection
Abstract:
Liquid biopsies are revolutionizing diagnostics in numerous ways. Unfortunately, RNA based liquid biopsies have proven difficult to validate as current methods of RNA detection are dependent on samples that exclude fragmented RNA. This despite reports that up to 90% of RNA in biofluids correspond to fragmented RNA. The RiboMarker technology addresses this limitation by allowing the efficient detection of fragmented RNA. RiboMarker’s enabling technology unlocks next-generation RNA analytes for broad diagnostic applications from liquid biopsies for oncology or infectious disease, to soil sampling. In this presentation we show data from a cohort of patients infected with Coccidioides (Valley Fever causal agent). Our data showcases how RiboMarker technology enables precise disease profiling using RNA fragmentomics, illustrating the practical implications of this innovation and its broader applicability in diagnostic settings.

Talk Title:
Advances in Biointelligence: From Multi-Omics to Multi-Source Fusion
Abstract:
In this talk, we will discuss how multi-omic data of the environmental microbiome is integrated with multi-source data and used in AI-powered analytics to increase biosecurity and public health. Current approaches to syndromic surveillance have fallen short in providing early warning for biological threats. With globally increasing biological risks due to disease spillover from animals, misuse, and other sources, the world needs a better way to detect, understand, and respond to these threats. Biological intelligence (BIOINT) provides this pathway. BIOINT represents a new paradigm that is persistent and pervasive, and transcends beyond the reactive approach of moving from crisis to crisis. BIOINT brings pathogen monitoring closer to where emergence occurs, and takes a layered approach integrating genomic data with other data sources. Technology is reducing the barriers to implementation, but urgent action is needed to create the infrastructure for collecting, analyzing, and operationalizing this powerful data.

Talk Title:
Microbial Culturomics as Part of the OMICS: an Essential Tool for Microbiome-Based Therapeutics
Abstract:
The advance and integration of omics technologies have profoundly transformed our understanding of the microbiome, significantly influencing the development of novel therapeutic strategies. Notably, microbial culturomics emerges as a critical component within this suite of technologies, reshaping our approach to microbiome-based therapies.
This presentation will examine the role of microbial culturing in general and particularly culturomics as part of the broader omics toolkit, emphasizing its transformative impact on the microbiome field. We will begin by assessing how multi-omics tools and approaches have enhanced our understanding of the microbiome, with a special focus on the value of microbial culturing methods and conditions. Despite the strides made by genome sequencing and metabolomic profiling, these techniques often neglect uncultured, low-abundance microbes and obscure metabolic pathways.
Microbial Culturomics bridges this gap by facilitating the isolation and characterization of previously known and unknown microbial taxa, thereby incrementing the microbial databases and synergizing with other omics strategies. Using culturomics we have built a collection of 1500+ bacterial isolates from companion animals including novel genera and species not detected by sequencing.
Moreover, culturing methods are a necessary step in Live Biotherapeutic Products (LBPs) development.Our perspective advocates for a cohesive multi-omics approach in microbiome research, addressing the significant gap between the estimated number of microorganisms and cultured microbes.

Talk Title:
Measurement Assurance for Innovation and Translation of Microbiome Science
Abstract:
Appreciation for the role of microbes in our lives has been growing rapidly, but the measurement science needed to understand and fully utilize microbial systems has developed at a much slower pace than the industries dependent on them demand. In all applications involving complex microbial communities, the research is hampered by the lack of standards, protocols, and technical infrastructure to allow confidence in the data and comparability. NIST is developing standards to enable research investigations and commercial translation of microbiome science by providing measurement assurance tools: protocols, reference materials, validated measurements, and critically evaluated reference data.

Talk Title:
The Respiratory Microbiome as a Source of Therapeutic Strains for Disease Prevention
Abstract:
Resilient Biotics is a platform company developing microbiome products to protect against respiratory infections and reduce the use of antibiotics in the livestock industry. Despite the routine use of vaccines and antibiotics, the respiratory disease burden remains high for cattle, swine, and poultry. Resilient Biotics has developed several microbiome therapeutic formulations employing both ecological and immunological mechanisms to address pathogens in the respiratory tract. Intranasal formulations have been validated in bovine, swine, and murine models. We are currently developing novel, multi-strain formulations that could be safely delivered in feed to reduce respiratory pathogen colonization and infection in nursery pigs. An in-house strain collection of more than 5,000 porcine respiratory colonizers has been constructed, and customized in vitro assays were employed to identify specific strains with pathogen inhibitory and immunostimulatory properties. Our products will enable new preventative strategies to combat respiratory pathogens in livestock.

Talk Title:
Multi-modal AI For Translational Research & Precision Medicine
Abstract:
At BioAI, we develop world-leading machine learning technology to develop digital biomarker tests for patient selection and screening. The BioAI PredictX platform is capable of ingesting a range of data types, including Digital Pathology, Multiomics and Real-World Evidence. Using multimodal data, state-of-the-art AI methods, and integrated deep learning, we can build predictive and prognostic models across a wide range of therapeutic areas. BioAI has built numerous models that can be used to classify both molecular status and tissue biomarkers directly from H&E slides without the need for additional molecular or IHC testing. PredictLung, currently in development, is an AI-powered digital test panel for NSCLC patient tumor tissue samples that can predict actionable mutations and biomarkers from H&E stained images. This test leverages an existing H&E image (available as a standard diagnostic procedure) and is a rapid, generalizable screening test that can help guide therapy selection.

Talk Title:
Microbiome 101: The Possibilities & Future of Clinical Microbiome Applications
Abstract:
Human microbiomes have become increasingly important in understanding many chronic diseases of the human condition. This talk aims to provide an overview of clinically important human microbiomes, the technologies used to characterize microbiomes, selected case studies illustrating the promise microbiome testing holds to improve human health, and discussion on the future of clinical microbiome testing.
%20Uzun_Headshot.jpeg)
Assistant Prof. of Pathology & Laboratory Medicine
The Warren Alpert Medical School of Brown University
Talk Title:
Unraveling Cancer: Identifying Drug Targets through Protein-Protein Interaction Networks
Abstract:
Patients diagnosed with the same cancer type can exhibit a wide array of genetic and physiological differences, including disparate responses to treatment, varied prognoses, and survival rates. In our study, we employed a novel method to study protein-protein interactions (PPIs) across 10 tumor types and their subtypes using publicly available genomic data from patients. Using this PPI data, we created a web-based application that displays PPI networks through interactive diagrams.

Talk Title:
Microbiome tRNA Sequencing and Application
Abstract:
The trillions of microbes that co-habit human bodies play a cardinal role in health and disease. It is paramount to fully identify, characterize and understand the relationships with and the effects of microbes on humans. We are investigating the relationship between microbes and humans in laboratory and clinical settings using a universally present cellular RNA family. Transfer RNAs (tRNAs) are required for protein synthesis and all cellular life. tRNAs also carry many epitranscriptomic modifications for their function. tRNA sequences are sufficiently distinct among prokaryotes and humans so that a single high throughput sequencing experiment can characterize tRNAs from both branches of life simultaneously. We have developed tRNA-seq approaches that overcome the difficulty of sequencing tRNA and their modifications. Here we discuss the application of microbiome tRNA-seq for laboratory and clinical studies.

Talk Title:
Microbiome in Aging
Abstract:
Join us for an enlightening webinar as Dr. Yadav, a world-renowned leader in microbiome and aging research, delves into the fascinating connection between gut health and brain aging. In this session, we will explore how a leaky gut, induced by microbiome imbalances, can contribute to cognitive decline and other age-related brain health issues, including multiomics data and outcomes from the MiaGB Consortium. Dr. Yadav will share cutting-edge technologies developed for collaborations, such as Postbiotics formulations and probiotics development platforms. Discover how anti-aging screenings using C. elegans and other innovative techniques are advancing our understanding of microbiome-brain interactions. Don’t miss this opportunity to learn from a trailblazer in the field and explore practical applications for healthier aging.

Talk Title:
Fueling Discovery Through Genomic and Metabolomic Analyses of Clinical Trials of Microbiome Therapeutics
Abstract:
Seres Therapeutics, a leading microbiome therapeutics company, has pioneered advancements in the field through clinical innovation and the integration of multi-omics data. Our journey began with foundational studies in microbial ecology and function, progressing to sophisticated clinical trials that employ metagenomic and metabolomic endpoints. A key challenge has been inferring bacterial function from complex microbiome data. To address this, we have utilized multi-omic datasets from our clinical interventional studies to inform predictive models of microbial function. This approach has led to significant breakthroughs, including advancements in identifying bacterial species that produce metabolites of therapeutic interest. Our work underscores the potential of continued advancement in microbiome therapeutics to treat a diverse array of conditions.

Talk Title:
A Gram-negative-selective antibiotic that spares the gut microbiome and prevents Clostridioides difficile infection
Abstract:
Infections caused by Gram-negative pathogens are increasingly prevalent and are typically treated with broad-spectrum antibiotics, resulting in disruption of the gut microbiome and susceptibility to secondary infections. There is a critical need for antibiotics that are selective both for Gram-negative bacteria over Gram-positive bacteria, as well as for pathogenic bacteria over commensal bacteria. Here we report the design and discovery of lolamicin, a Gram-negative-specific antibiotic targeting the lipoprotein transport system. Lolamicin has activity against a panel of more than 130 multidrug-resistant clinical isolates, shows efficacy in multiple mouse models of acute pneumonia and septicaemia infection, and spares the gut microbiome in mice, preventing secondary infection with Clostridioides difficile. The selective killing of pathogenic Gram-negative bacteria by lolamicin is a consequence of low sequence homology for the target in pathogenic bacteria versus commensals; this doubly selective strategy can be a blueprint for the development of other microbiome-sparing antibiotics.
Conference Sponsors
VIRAL SPONSORS



BACTERIAL SPONSORS
FUNGAL SPONSORS



SPEAKERS

Cargill

National Institute of Standards & Technology
NIST



Agenda Day 1
Wednesday June 21st

Zachary Pitluk, PhD
VP Life Science BD
Paradigm4
Talk Title: Uniting Kingdoms - How to Manage Omics Data from Helicobacter to Humans using REVEAL: SingleCell
Abstract:
While the term meta-organism has been used to describe the obligate nature of prokaryotes for mammalian survival, the relationship has always been at flagella's length. With the increasingly central role that the human microbiome plays, efforts focused on truly integrating data to support joint discovery across kingdom boundaries. Challenges, like managing sparsity of data and the scale of the feature maps, can be solved using modern data modeling and management techniques, like those that are in Paradigm4's REVEAL platform.

Kristin Neumann, PhD
Co-Founder & CEO
MyMicrobiome
Talk Title: What the Bleep Do We Know about the Microbiome?
Abstract:
The microbiome is an exciting new area humanity is exploring. After the decoding of the human genome, we realized that our genome does not offer sufficient tools to make our body work properly. This is how the Human Microbiome Project was initiated. Now, 16 years later, the microbiome became mainstream. Products analysing, supporting and healing our microbiome are flooding the market, spreading the hope to improve everyone’s health.
What do we currently know about this amazing topic and how can applications be used in a meaningful way? In this talk we are going to summarize the current state of research and what can be done to support a healthy microbiome in our lives.

Alexander Senf, PhD
Senior Bioinformatics Engineer
Biomatics
Talk Title: Beyond Genomics: Multi-Omics and Microbiome Data and the Importance of FAIRness
Abstract:
An overview of omics data beyond genomic sequences and sequence variations, leading to the importance of data being FAIR. The microbiome is introduced, in terms of omics data, leading to a case study underlining the usefulness of FAIR data when analyzing data in this environment.

Syed Ana Jawaid
Bioinformatics Analyst
Biomatics
Talk Title:
Abstract:

Ken Finley, PhD
Principal Biotechnologist
Cargill
Talk Title: Galleon - Cargill’s Microbiome Intelligence Tool
Abstract:
Cargill’s Galleon™ Microbiome Intelligence is a simple, non-invasive analysis tool that allows producers to determine the gut microbiome health of their flocks. After sampling, poultry producers receive a report that includes a summary of their site samples, flock microbiome biomarker composition, predictions and insights about the microbiome constitution of their flock. A Cargill technical expert walks producers through the report in detail and provides recommendations on how to take action to support performance, health and preharvest food safety. Galleon™ simplifies a complex process to provide poultry producers with practical, actionable insights to help improve animal health, performance, food safety, and ultimately, a return on investment through non-invasive testing of the gut microbiome health of their flock. Cargill’s Galleon™ uses an AI-powered engine that allows producers to determine the gut microbiome fitness of their flock while also helping them understand how it correlates to numerous management parameters and practices.

Pedro Torres, PhD
Sr. Scientist, Computational Biology & Data Science
Persephone Biosciences
Talk Title:
My Baby Biome Study: A Data-Driven Approach to Infant Probiotic Development
Abstract:
The infant microbiome plays a crucial role in establishing a healthy host-microbiome relationship. Recent research has identified a critical window in early-life (the first 3 months) where gut microbial dysbiosis has the most impact on human immune development. Existing infant microbiome datasets lack comprehensive multi-omics analysis and sufficient racial and ethnic diversity for population-wide biomarker discovery. Persephone Biosciences addressed this gap through the My Baby Biome (MBB) study, enrolled ~700 infants up to the age of 3 months across the U.S, representative of US birth and feeding modes Using shotgun metagenomic sequencing and metabolomics, we identified three microbiome compositional clusters. A healthy cluster was distinguished by high Bifidobacterium levels and association with specific anti-inflammatory metabolites. Further exploration of MBB isolated strains and commercial strains in ex-vivo gut environments revealed their impact on microbial composition and function. These findings provide a data-driven approach to infant probiotic development.

Eddie Adams, PhD
Chief Scientific Officer
Micronoma
Talk Title:
Using All Our Genomes: Multi-Species & Multi-Omic Liquid Biopsy Detection of Early-Stage Lung Cancer
Abstract:
Links between cancer and microbes date back four millennia (Sepich-Poore et al. 2021. Science). Recently, we found that microbial DNA is detectable in tumor tissues and patient blood from many human cancer types (Poore et al. 2020. Nature; Narunsky-Haziza et al. 2022. Cell). These intratumoral and bloodborne microbiomes were distinct between cancer types, between normal and malignant tissues, and present in cell-free plasma samples. However, the practical utility of cell-free microbial DNA (cf-mbDNA) as a bona fide liquid biopsy remains untested, including its ability to diagnose early-stage disease in treatment-naïve individuals, to distinguish histological subtypes, and to discriminate against non-cancer-but-diseased patients. Thus, we constructed an age and sex-matched cohort (“CODICES”) comprising >1000 individuals with lung cancer, lung disease, and no disease (healthy) to evaluate the utility of a cf-mbDNA-driven liquid biopsy diagnostic. We show that cf-mbDNA comprises a novel class of cancer-genome orthogonal biomarkers that complements host analytes.

Joseph Petrosino, PhD
Director, Alkek Center for Metagenomics & Microbiome Research & Professor
Baylor College of Medicine
Talk Title: Translational Microbiome Research in the Era of Personalized Medicine
Abstract:
The Alkek Center for Metagenomics and Microbiome Research is coordinating and leading research and development efforts in this area across the Texas Medical Center, and with collaborators from around the US and abroad. CMMR researchers are developing molecular and informatics tools and resources to advance diverse clinical and basic research projects pertaining to the organisms that comprise the microbiome, the genetic makeup of these bacteria, viruses and fungi, and how these commensal microorganisms interact with human cells and tissues during the course of life. In addition to germ free animal models, C. elegans and zebrafish, we are leveraging sophisticated 3D tissue culture systems to understand the reveal personalized interactions between the microbiome and host. These tools have helped us reveal causal microbiome interactions that may be critical for human health and development.

Kuan Ding, PhD
Chief Science Officer
Catalytic Data Science
Talk Title: In silico Hypothesis Generation and Testing for Microbiome Research
Abstract:
In recent years, the study of microbiomes and multiomics data has gained immense importance in unraveling complex biological systems. The integration of the various omics data sets, such as genomics, transcriptomics, metabolomics, and proteomics, with microbiome data has paved the way for a deeper understanding of the interplay between microbial communities and host health. However, extracting meaningful insights from the vast amounts of data generated by these technologies remains a significant challenge.
This presentation aims to highlight the power of Catalytic Data Science in advancing microbiome research with an approach to accelerate hypothesis generation with advanced, integrated multiomic data analysis. We showcase real-world applications of Catalytic Data Science in Crohn’s Disease, including an end to end workflow identifying candidate genes, pathway associations, and subsequent downstream analyses.
Catalytic Data Science has developed a comprehensive, cloud-based solution designed to empower life sciences researchers to streamline their data management, analysis and collaboration efforts. Catalytic Data Science combines machine learning, statistical modeling, and domain expertise with advanced analytics and visualizations to extract actionable insights from complex biological datasets.

Scott Peterson, PhD
VP Translational & Laboratory Science
Prescient Metabiomics
Talk Title:
Meta-analysis of Multi-functional Biomarkers for Predictive Modeling of Colorectal Adenoma and Carcinoma
Abstract:
We conducted a clinical meta-analysis of published deep metagenomic stool sequence datasets including 1,670 subjects from 9 countries, including 703 healthy controls, 161 precancerous colorectal adenoma (CRA), 48 advanced precancerous colorectal adenoma (CRAA) and 758 CRC cases confirmed by colonoscopy. We describe a novel automated machine learning workflow using a two-stage feature importance ranking and ensemble modeling method to identify and select highly predictive taxonomic and functional biomarkers. Machine learning modeling of selected features differentiated the metagenomic profiles of healthy patients from CRA, CRAA and CRC cases with an average area under the curve (AUC) for external holdout testing of 0.84 (sensitivity=0.82; specificity=0.71, accuracy=0.77) for CRC; an AUC of 0.97 (sensitivity=0.78; specificity=0.98, accuracy=0.97) for CRAA; and an AUC of 0.90 (sensitivity=0.74, specificity=0.89, accuracy=0.86) for CRA. Application of our ensemble approach for feature selection increased the predictive power and may generate greater generalizability.

Stephanie Servetas, PhD
Microbiologist
National Institute of Standards & Technology (NIST)
Talk Title:
Measurement Assurance for Innovation in Microbiome Science
Abstract:
Appreciation for the role of microbes in our lives has been growing rapidly, but the measurement science needed to understand and fully harness microbial systems has developed at a much slower pace than the industries dependent on them demands. In all applications involving complex microbial communities, the research is hampered by the lack of standards, protocols, and technical infrastructure to allow confidence in the data and comparability. At NIST, we are developing tools to enable measurement assurance of complex microbial systems for applications in clinical diagnostics, agriculture, and the environment.

Keith Robison, PhD
Principal Scientist
Ginkgo Bioworks
Talk Title:
Let's Do The Warp Drive Again! Can Sequencing Exhaustively Elucidate the Microbiome?
Abstract:
A bit over a decade ago, Warp Drive Bio formed to discover transformative natural product pharmaceuticals. In its six-year lifespan, WDB generated the largest industrial microbe database (about 145K genomes) for this purpose, yet multiple datapoints suggest it barely scratched the surface of the biochemical diversity of the Actinomycetes. Sequencing technology has improved immensely in the intervening time - if we tried again how much better could we expect to do? Are the new sequencing instruments up to the task? What are the gaps in DNA extraction and library prep technology that have resulted from the industry's focus on human not microbe genomes?

Vancheswaran Gopalakrishnan, PhD
Associate Director, Bioinformatics
AstraZeneca
Talk Title:
Abstract:

Casandra Hoffman
Director, Research Affinity Groups
University of Virginia
Talk Title:
Supporting Microbiome Research at Universities: UVA’s TransUniversity Microbiome Initiative
Abstract:
The infant microbiome plays a crucial role in establishing a healthy host-microbiome relationship. Recent research has identified a critical window in early-life (the first 3 months) where gut microbial dysbiosis has the most impact on human immune development. Existing infant microbiome datasets lack comprehensive multi-omics analysis and sufficient racial and ethnic diversity for population-wide biomarker discovery. Persephone Biosciences addressed this gap through the My Baby Biome (MBB) study, enrolled ~700 infants up to the age of 3 months across the U.S, representative of US birth and feeding modes Using shotgun metagenomic sequencing and metabolomics, we identified three microbiome compositional clusters. A healthy cluster was distinguished by high Bifidobacterium levels and association with specific anti-inflammatory metabolites. Further exploration of MBB isolated strains and commercial strains in ex-vivo gut environments revealed their impact on microbial composition and function. These findings provide a data-driven approach to infant probiotic development.
Agenda Day 2
Thursday, June 22nd

Simon Adar, PhD
CEO
Code Ocean
Talk Title:
Abstract:
Announcing The World’s First
Microbiome & Multi-Omics (M&M)
Virtual Conference
March 8-9, 2022
The amount of work happening today to understand biology has and will continue to exponentially increase. What comes along with that is exponential increases in data!
We will get to see major discoveries unfold over the next couple of years because of the microbiome and multi-omics research. We predict we will also see many associations and improvements in all aspects of life simply through the new understanding and how the microbiome is involved in multiple different systems.
Central to science and technology endeavors and understandings is and will be the ability to produce and consume FAIR data and processes and use the power of technology to exploit as much as possible.
Because of these immense current and future capabilities, 20/15 Visioneers will bring together the thought leaders who are making major impacts from a software, research, and product perspective. In the spirit of keeping things simple, highly interactive, and focused on knowledge sharing the live virtual conference will consist of the following:
Pivoting the typical conference approach while reducing the tracks and bringing the most interesting presentations for a better attendee experience.
-
Daily Round Table summaries
-
Key Notes
-
Vendor Show Cases with individual Break-Out-Rooms (Augmented Reality), scheduled 2x each day
-
Industry and academic speakers
-
Free for attendees!
Conference Recording Links
.jpeg)















